2 Package Development
In R, the fundamental unit of shareable code is the package. A package bundles together code, data, documentation, and tests, and is easy to share with others.
But packages are useful even if you never share your code. As Hilary Parker says in her introduction to packages: “Seriously, it doesn’t have to be about sharing your code (although that is an added benefit!). It is about saving yourself time.” Organising code in a package makes your life easier because packages come with conventions. For example, you put R code in R/, you put tests in tests/ and you put data in data/. These conventions are helpful because:
They save time — you don’t need to think about the best way to organise a project, you can just follow a template.
Standardised conventions lead to standardised tools — if you buy into R’s package conventions, you get many tools for free.
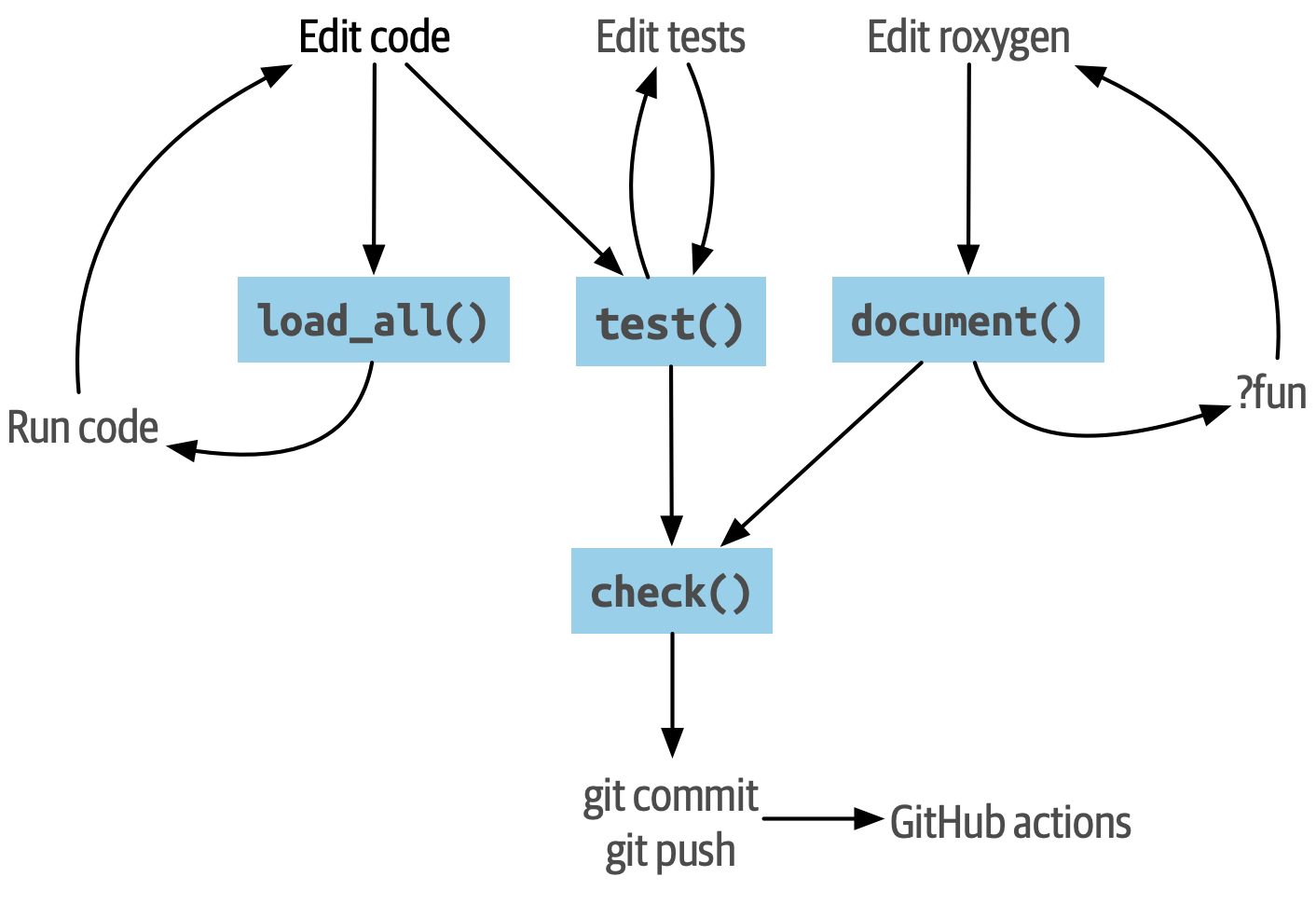
2.1 Package Structure
In an R package or R project structured as a package the typical files and folders will be (locally, you can consult your Files pane):
| path | type | description |
|---|---|---|
| .Rbuildignore | file | Lists files that we need to have around but that should not be included when building the R package from source. |
| .gitignore | file | Tells Git to ignore some standard, behind-the-scenes files created by R and RStudio. |
| DESCRIPTION | file | Provides metadata about your package. |
| NAMESPACE | file | Declares the functions your package exports for external use and the external functions your package imports from other packages. |
| R/ | folder | the “business end” of your package. It will soon contain .R files with function definitions. |
2.2 Loading devtools and usethis
The devtools package is fundamental for developing packages, it comes with a suite of incredibly powerful functions. In addition,
it comes with the required package usethis, which compliments the devtools package with another suite of functions required to properly build packages.
2.3 load_all function
In a package or project structured as a package you are typically making functions that are stored in the R/ folder. In a standard project you may be familiar with the use of source("R/myfunction.R") to load or run a script. However, devtools allows us to easily run/load all of our project contents with one simple function call:
This does a few main things:
- Loads/runs your scripts located in the
R/folder
- Loads data stored in your
datafolder
- Loads other package objects
- Loads package dependencies listed in the
DESCRIPTIONfile
NOTE
One main difference is that these functions and data that have been loaded will not appear in the environment, even though they are available. This is similar to when we load a package, such as library(tidyverse), we are now able to use functions such as mutate even though they don’t appear in our environment.
2.4 The DESCRIPTION file
The DESCRIPTION file provides metadata for your package. Some key pieces of this metadata include the description of the project and the dependencies.
If your project doesn’t have a DESCRIPTION file you can easily add one using usethis:
You can manually edit this file or alternatively add certain elements using usethis. For example adding dependencies:
NOTE
After creating a DESCRIPTION file in your project you will automatically enter package development mode.
2.5 Documenting your functions
At some point we have all used the help functions in R by easily calling something like ?mutate. This requires special documentation which is stored in path such as man/mutate within the package. To do this for ourselves we have to use something called roxygen2, which helps create these handy help windows. To do this with your functions you can open your function script, place the cursor somewhere within the function and then do Code > Insert roxygen skeleton, which will create a basic skeleton to fill out such as this:
#' Split a string
#'
#' @param x A character vector with one element.
#' @param split What to split on.
#'
#' @return A character vector.
#' @export
#'
#' @examples
#' x <- "alfa,bravo,charlie,delta"
#' strsplit1(x, split = ",")
strsplit1 <- function(x, split) {
strsplit(x, split = split)[[1]]
}Now, one more step is needed. We must use devtools to automatically create that man/function and update our NAMESPACE file like so:
2.6 The NAMESPACE
The NAMESPACE file is an automatically generated and maintaind file by R, this should not be manually modified. It is filled out depending on the roxygen2 comments left in your scripts and is updated, as described above, by using devtools::document(). It informs the package what contents should be exported when building the package, as well as what needs to be imported (package dependencies) for the package to run.
2.7 The README file
The README file is a very useful document that can help provide context, general information, and usage insight to users. In addition, when knitted, README files are formatted to appear as nice markdown documents in Github and Gitlab.
To get a README file started in a project all that you need to do is:
NOTE
Remember, you have to knit your README in order to produce a .md file version of it, which will be directly used in places like Github or Gitlab.
2.8 Organizing your scripts
The file name should be meaningful and convey which functions are defined within. While you’re free to arrange functions into files as you wish, the two extremes are bad: don’t put all functions into one file and don’t put each function into its own separate file.
| Organizing.principle | Comments |
|---|---|
| One function | Defines exactly one function, that’s not particulary large, but doesn’t fit naturally into any other .R file |
| Main function plus helpers | Defines the user-facing function, a method, and private helpers |
| Family of functions | Defines a family of functions, all documented together in a big help topic, plus private helpers |
TIP
Another file you often see in the wild is R/utils.R.
This is a common place to define small utilities that are used inside multiple package functions.
Since they serve as helpers to multiple functions, placing them in R/utils.R makes them easier to re-discover when you return to your package after a long break.
2.9 Using data in a package
Traditionally, data in a package is stored in the data/ folder. The data there will be saved in a specific data form that will make it available when you run devtools::load_all(). To store data within a package like this you need to run: